UNDER CONSTRUCTION!! Please email alanim@pennmedicine.upenn.edu or sgourakisn@chop.edu for job submissions while we fix an error with the frontend server. MAUS (Methyl Assignments Using Satisfiability) is a computational method which performs reliable assignments of methyl NMR peaks observed in 2D HMQC 13C/1H correlation spectra, to methyl groups in a protein of interest. To do this, MAUS makes use of through-space connectivities present in 3D or 4D NOESY spectra and a 3D structural model of the protein that can be obtained from the Protein Data Bank (rcsb.org), or using comparative modeling (Song et al.) and sequence co-evolution approaches (Ovchinnikov et al.) or through deep learning approaches (Jumper et al.).
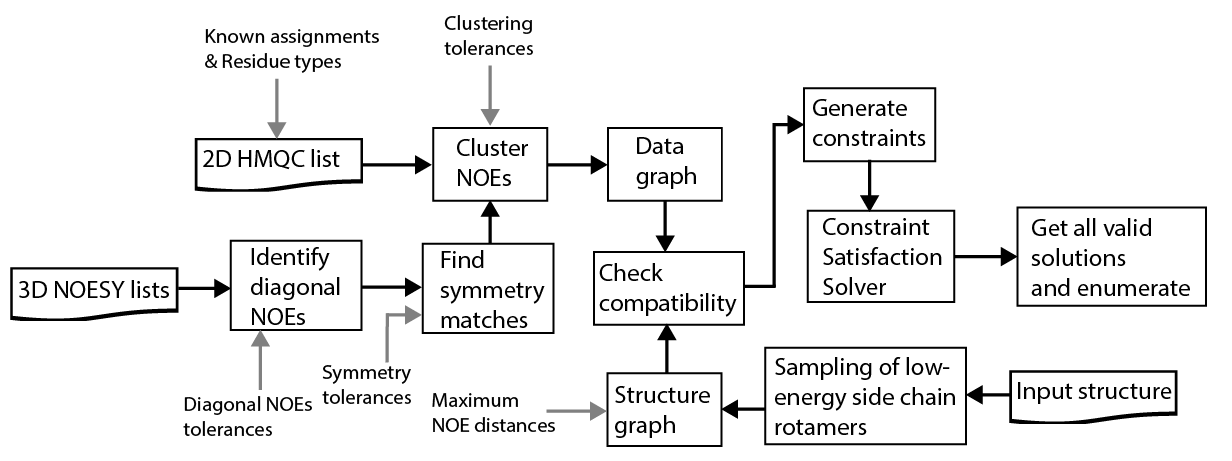
Assigning 2D NMR peaks to methyl groups in a protein structure using a network of NOEs is a computationally intractable problem. The complexity of this problem arises from the astronomically large (10140) number of possible assignments for a typical 20-30 kDa size protein molecule.
However, not all solutions are valid: Under the assumption that the NOE effect probes distances of up to 10-12 Å, the number of valid assignments is reduced by 100 orders of magnitude (Pritišanac et al.). Similarly, there are additional considerations that can be drawn from the input data which allow elimination of large solution sub-spaces.
We treat these rules as hard constraints and give them as inputs to a custom-made Constraint Satisfaction Solver. Using a fast iterative process, the solver then enumerates all valid assignments for each 2D methyl peak (see schematic below).
We find that the information in the input NOE data is sufficient to provide unambiguous assignments for a large fraction (up to 90%) of methyl peaks. The results are presented to the user in the form of a comprehensive table, which allows her to go back to the original NOESY spectra and eliminate assignments that are inconsistent with the observed NOE peak intensities, not considered by MAUS.
Nerli, S., De Paula, V.S., McShan, A.C. et al. Backbone-independent NMR resonance assignments of methyl probes in large proteins. Nat Commun 12, 691 (2021). https://doi.org/10.1038/s41467-021-20984-0
Patent pending. Free unlimited use for academic purposes.
If you encounter any errors, please email sagarg@sas.upenn.edu and sgourakisn@chop.edu